MolProbity Ramachandran analysis
Transcrição
MolProbity Ramachandran analysis 3P9D, model 1 General case 180 E M11 462 462 B JARG ARG 202 202 LYS LYS C 52 MET IAB 11 ASP ASP JK 142 142 ASN ASN P H 322 LYS P H 322 122 122LYS LEU LEU K C 16 THR K CF N 321 353 353LYS LEU LEU F N 343 343 LEU LEU F N 167 167 THR THR B JGLN 246 246 IA 508 508 PHE PHE LYS LYS C 265 LYS P H 373 373K GLN IAE M265 265 438 438ASP ASP ARG ARG P HB J 343 343 327 327ARG SER ARG SER B J 307 30713 GLU GLU P H 13 LEU LEU 518 518ASN LEU LEU L D G OIA188 188 362 ASN TYR G O 224 250 LEU E M 376 376 THR THR G O SER 479 IA 367 367 SERTYR G O PHE IA 337 LEU LEU K C 337 G O 258 256 GLN ASNN F 486 THR THR G O 337 THR N 49 467 467 LEU LEU P HF 49 CYS CYS E M 369 369 LEU LEU Psi Isoleucine and valine 180 H 467 378 378ILE ILE ILE B JP 467 ILE G O 62 ILE Psi P H 150 150 ILE ILE B J 35 35 SER SER IA 55 55ASP ASP L D 99 ALA ALA C 67 36 ARG G OK P H 187 187ALA SER SER L D 95 SER F N 95 24 24SER THR THR 0 O190 495 PHE E MG 190 SER SER B J 184 184 LEU LEU IA 56 56ASP ASP K CJB 252 370 370TYR ARG ARG E M 507 507 ILE ILE P H 484 484 VAL F N VAL 138 138 ILE ILE F N 489 489 VAL VAL P H 147 147 VAL VAL 0 E MG 402 402 LYS LYS O ASP B J 301 301190 LEU LEU F N 188 188 ASN ASN IA 74 74ASP ASP B J 478 478 THR THR F N 183 183 ASP ASP E M 351 351 LEU LEU P H 483 483 ASN ASN B J 372 372 ALA ALA L D 431 431 SER SER B JALA 16 16 GLU GLU K CG 30 M 147 147 ALA O 428 SER K C 256 GLU P HK CE 356 P H 356 460 178 178 GLU GLU ALA LEU LEU F N 298 298 GLN GLN B JK C454 144 50 SER SER ASP L DK L D382 382 485 485 K CF ALA ALA 19 ARG ARG GLN C ARG N50 460 460 LYS LYS L D 427 427 LYS LYS L D 294 294 ARG ARG K C 303 ASP G O 487 ASN L D 474 474 GLU GLU -180 IA 94 94ILE ILE G O 63 SER B J 220 220 ASN ASN J 470 470 SER SER F N 160 160 THR THR B K C 490 LYS 0 Phi P H 490 490 VAL VAL B J 55 ILE ILE -180 0 Phi M 254 254 MET MET E IA 77 77HIS HIS 121 121 HIS HIS G O IA368 CYS 180 Glycine 180 E M 251 251 HIS HIS E M 440 440 VAL VAL -180 180 Pre-proline 180 F NP 512 512 VAL VAL P H 284 284 ILE ILE 146 VAL 146 VAL HH502 458 458 VAL VAL OP B J 88 88G VAL VAL VAL E M 167 167 ASP ASP J 15 15 ALA ALA B J 185 185 LYS LYSB L D 205 205 ASP ASP G O 478 TRP ID A 335 335 ALA ALA O 213 LEU L 143 143 SER SER P HG 233 233 GLU GLU -180 B J 165 165 ILE ILE IA 376 376GLY GLY B J E M 323 323 386 386 GLY GLY GLY GLY F N 412 412 ILE ILE M E 347 347 GLY GLY Psi Psi 0 0 F N 333 333 GLY GLY E M 36 36 GLY GLY B J 371 371 GLY GLY G O 45 GLY P H 486 486 GLU GLU B J 293 293 TYR TYR G OE M528 493 493 GLY GLY GLY IA 292 292GLY GLY IA 22 22GLY GLY N 490 490 GLY GLY G O 412 GLY F -180 K C 50 ASP -180 -180 E M 274 274 CYS CYS 0 Phi 180 Trans proline 180 -180 0 Phi 180 Phi 180 Cis proline 180 IA 267 267PRO PRO K C 249 PRO K C 142 PRO E M IA 199 279 199 279PRO PRO PRO PRO Psi Psi K C 410 PRO 0 0 F N 204 204 PRO PRO B J 117 117 PRO PRO P H 460 460 48 48 PRO PRO PRO PRO L D 17 17 PRO PRO G O 210 46 46 PRO PRO K C PRO IAP 549 PRO PRO H549 324 324 PRO PRO P H 66 PRO PRO -180 -180 -180 82.9% (6885/8302) of all residues were in favored (98%) regions. 96.2% (7986/8302) of all residues were in allowed (>99.8%) regions. There were 316 outliers (phi, psi): A 11 ASP (47.8, 176.5) A 22 GLY (43.7, -104.7) A 55 ASP (-71.5, 30.8) A 56 ASP (38.4, 25.0) A 74 ASP (100.2, -16.2) A 77 HIS (92.8, 152.0) A 94 ILE (-148.3, -91.7) A 121 HIS (79.1, 126.5) A 199 PRO (-38.6, 103.6) A 265 ASP (160.6, 146.6) A 267 PRO (-40.3, 160.8) A 292 GLY (176.9, -81.6) A 335 ALA (-60.7, -171.9) A 337 LEU (-54.9, 96.6) A 367 SER (64.8, 98.4) A 376 GLY (67.0, 142.1) A 508 LYS (106.3, 153.8) A 518 LEU (176.6, 122.5) A 549 PRO (-75.4, -156.8) B 5 ILE (81.3, -175.7) B 15 ALA (79.5, -143.3) B 16 GLU (-25.4, -71.1) B 35 SER (-62.2, 52.8) B 50 SER (-52.1, -82.7) B 88 VAL (-82.6, -84.8) B 117 PRO (-77.0, -54.7) B 142 ASN (50.6, 173.2) B 165 ILE (-18.4, -39.2) B 184 LEU (100.7, 31.5) B 185 LYS (32.3, -145.3) B 202 LYS (86.8, 179.6) B 220 ASN (84.8, -163.8) B 246 PHE (92.8, 153.1) B 293 TYR (-110.5, -59.0) B 301 LEU (99.5, 0.3) B 307 GLU (52.6, 124.7) B 323 GLY (63.5, 132.8) B 327 SER (67.3, 140.6) B 370 ARG (178.1, 38.1) B 371 GLY (-25.1, -42.4) B 372 ALA (-26.4, -51.9) B 467 ILE (60.4, 115.1) B 470 SER (159.6, -166.8) B 478 THR (101.8, -20.0) C 16 THR (-14.2, 147.7) C 19 GLN (-45.4, -83.3) C 30 ALA (-52.1, -73.2) C 36 ARG (-63.2, 18.2) C 50 ASP (-61.4, -175.7) C 52 MET (54.1, 177.8) C 142 PRO (-99.0, 115.2) C 144 ASP (-51.2, -82.2) C 210 PRO (-45.7, -97.8) C 249 PRO (-111.4, 131.9) C 252 TYR (174.3, 39.5) C 256 GLU (-47.0, -77.7) C 258 GLN (-53.5, 95.6) C 265 LYS (92.5, 151.7) C 303 ASP (93.3, -122.9) C 321 LYS (-25.5, 140.3) C 410 PRO (-61.3, 11.7) C 454 ARG (-61.9, -85.6) C 460 ALA (-56.8, -79.0) C 490 LYS (49.3, -173.9) D 9 ALA (-69.3, 23.9) D 17 PRO (-44.9, -80.9) D 95 SER (-54.3, -2.6) D 143 SER (-60.8, -173.1) D 188 ASN (170.9, 119.8) D 205 ASP (-64.0, -147.3) D 294 ARG (67.2, -95.5) D 382 ALA (-68.1, -83.4) D 427 LYS (-65.8, -94.0) D 431 SER (-23.4, -60.7) D 474 GLU (-177.4, -145.9) D 485 ARG (-60.4, -83.8) E 36 GLY (172.5, 26.2) E 147 ALA (-54.7, -75.0) E 167 ASP (149.1, -131.2) E 190 SER (86.7, 37.3) E 251 HIS (48.4, 176.8) E 254 MET (94.1, 157.0) E 274 CYS (85.8, -178.2) E 279 PRO (-38.6, 103.4) E 347 GLY (-16.9, 118.7) E 351 LEU (-18.4, -27.8) E 369 LEU (-25.4, 77.1) E 376 THR (-55.5, 100.8) E 386 GLY (71.0, 132.3) E 402 LYS (102.7, 5.9) E 438 ARG (163.8, 146.1) E 440 VAL (-85.8, -159.6) E 462 ARG (55.0, 179.7) E 493 GLY (-50.7, -80.5) E 507 ILE (40.0, 28.5) F 24 THR (-53.6, -4.5) F 138 ILE (82.6, 11.8) F 160 THR (94.5, -169.6) F 167 THR (167.4, 173.4) F 183 ASP (-43.9, -21.9) F 188 ASN (99.5, -8.7) F 204 PRO (-40.4, -9.9) F 298 GLN (-47.1, -81.5) F 333 GLY (17.3, 91.2) F 343 LEU (-24.7, 129.7) F 353 LEU (-20.7, 138.9) F 412 ILE (172.6, 131.8) F 460 LYS (-43.5, -86.0) F 467 LEU (-57.2, 82.8) F 486 THR (11.0, 92.4) F 489 VAL (92.3, 0.9) F 490 GLY (-10.3, -145.0) F 512 VAL (-58.2, -71.7) G 45 GLY (-19.1, -57.0) G 46 PRO (-45.1, -95.0) G 62 ILE (-165.4, 117.4) G 63 SER (176.2, -159.3) G 67 ALA (-66.6, 13.9) G 190 ASP (111.8, 4.3) G 213 LEU (-52.5, -172.9) G 224 PHE (-42.7, 97.1) G 250 LEU (-37.1, 101.0) G 256 ASN (-42.9, 95.5) G 337 THR (58.7, 91.9) G 362 TYR (173.0, 119.2) G 368 CYS (70.2, 122.1) G 412 GLY (-65.8, -148.9) G 428 SER (-49.7, -75.5) G 478 TRP (-59.8, -165.4) G 479 TYR (82.8, 99.1) G 487 ASN (-178.9, -123.7) G 495 PHE (96.3, 41.7) G 502 VAL (-58.1, -80.8) G 528 GLY (-58.6, -80.8) H 6 PRO (-56.9, -166.7) H 13 LEU (64.9, 122.7) H 48 PRO (-58.1, -74.7) H 49 CYS (-64.9, 78.8) H 122 LEU (-48.4, 167.6) H 146 VAL (-48.4, -76.0) H 147 VAL (63.5, -13.2) H 150 ILE (-61.3, 85.2) H 178 LEU (-46.7, -78.4) H 187 SER (-62.9, 9.2) H 233 GLU (-58.4, -176.5) H 284 ILE (-47.4, -72.2) H 322 LYS (-50.2, 168.7) H 324 PRO (-69.5, -160.1) H 343 ARG (63.9, 140.2) H 356 GLU (-59.1, -78.8) H 373 GLN (62.1, 150.8) H 378 ILE (62.8, 119.0) H 458 VAL (-55.2, -77.3) H 460 PRO (-58.4, -74.2) H 483 ASN (8.4, -40.2) H 484 VAL (60.7, 13.0) 0 Phi 180 -180 0 H 486 GLU (-144.2, -54.8) H 490 VAL (-162.4, -151.6) I 11 ASP (47.8, 176.5) I 22 GLY (43.7, -104.7) I 55 ASP (-71.6, 30.8) I 56 ASP (38.4, 25.1) I 74 ASP (100.1, -16.2) I 77 HIS (92.8, 151.9) I 94 ILE (-148.2, -91.7) I 121 HIS (79.1, 126.5) I 199 PRO (-38.6, 103.7) I 265 ASP (160.6, 146.7) I 267 PRO (-40.3, 160.8) I 292 GLY (176.9, -81.6) I 335 ALA (-60.7, -172.0) I 337 LEU (-54.8, 96.6) I 367 SER (64.8, 98.4) I 376 GLY (67.0, 142.1) I 508 LYS (106.3, 153.8) I 518 LEU (176.6, 122.5) I 549 PRO (-75.3, -156.8) J 5 ILE (81.3, -175.7) J 15 ALA (79.6, -143.3) J 16 GLU (-25.5, -71.1) J 35 SER (-62.3, 52.7) J 50 SER (-52.0, -82.7) J 88 VAL (-82.6, -84.8) J 117 PRO (-76.9, -54.7) J 142 ASN (50.6, 173.2) J 165 ILE (-18.5, -39.1) J 184 LEU (100.6, 31.5) J 185 LYS (32.3, -145.3) J 202 LYS (86.7, 179.7) J 220 ASN (84.8, -163.9) J 246 PHE (92.9, 153.1) J 293 TYR (-110.5, -58.9) J 301 LEU (99.5, 0.3) J 307 GLU (52.7, 124.7) J 323 GLY (63.5, 132.9) J 327 SER (67.3, 140.6) J 370 ARG (178.0, 38.2) J 371 GLY (-25.1, -42.4) J 372 ALA (-26.3, -52.0) J 467 ILE (60.5, 115.0) J 470 SER (159.6, -166.8) J 478 THR (101.8, -20.0) K 16 THR (-14.2, 147.7) K 19 GLN (-45.3, -83.3) K 30 ALA (-52.1, -73.2) K 36 ARG (-63.2, 18.2) K 50 ASP (-61.3, -175.7) K 52 MET (54.1, 177.8) K 142 PRO (-99.0, 115.2) K 144 ASP (-51.1, -82.1) K 210 PRO (-45.7, -97.8) K 249 PRO (-111.4, 131.9) K 252 TYR (174.3, 39.5) K 256 GLU (-46.9, -77.7) K 258 GLN (-53.5, 95.7) K 265 LYS (92.6, 151.7) K 303 ASP (93.2, -122.9) K 321 LYS (-25.4, 140.3) K 410 PRO (-61.4, 11.7) K 454 ARG (-61.8, -85.6) K 460 ALA (-56.9, -78.9) K 490 LYS (49.2, -173.9) L 9 ALA (-69.3, 23.9) L 17 PRO (-44.9, -80.9) L 95 SER (-54.3, -2.6) L 143 SER (-60.9, -173.1) L 188 ASN (171.0, 119.9) L 205 ASP (-64.1, -147.3) L 294 ARG (67.2, -95.5) L 382 ALA (-68.2, -83.4) L 427 LYS (-65.8, -94.1) L 431 SER (-23.5, -60.6) L 474 GLU (-177.4, -145.9) L 485 ARG (-60.4, -83.8) M 36 GLY (172.5, 26.2) M 147 ALA (-54.7, -74.9) M 167 ASP (149.1, -131.2) M 190 SER (86.6, 37.3) M 251 HIS (48.4, 176.7) M 254 MET (94.0, 157.0) M 274 CYS (85.9, -178.2) M 279 PRO (-38.6, 103.4) M 347 GLY (-17.1, 118.8) M 351 LEU (-18.4, -27.8) M 369 LEU (-25.4, 77.1) M 376 THR (-55.4, 100.7) M 386 GLY (71.0, 132.3) M 402 LYS (102.8, 5.8) M 438 ARG (163.8, 146.1) M 440 VAL (-85.9, -159.5) M 462 ARG (55.0, 179.7) M 493 GLY (-50.6, -80.5) M 507 ILE (39.9, 28.5) N 24 THR (-53.6, -4.4) N 138 ILE (82.7, 11.8) N 160 THR (94.5, -169.7) N 167 THR (167.3, 173.4) N 183 ASP (-43.9, -21.9) N 188 ASN (99.5, -8.6) N 204 PRO (-40.4, -10.0) N 298 GLN (-47.2, -81.5) N 333 GLY (17.3, 91.3) N 343 LEU (-24.7, 129.6) N 353 LEU (-20.6, 138.9) N 412 ILE (172.6, 131.8) N 460 LYS (-43.4, -86.0) N 467 LEU (-57.1, 82.8) N 486 THR (11.0, 92.4) N 489 VAL (92.3, 1.0) N 490 GLY (-10.2, -145.1) N 512 VAL (-58.2, -71.7) O 45 GLY (-19.2, -57.0) O 46 PRO (-44.9, -95.1) O 62 ILE (-165.4, 117.3) O 63 SER (176.2, -159.3) O 67 ALA (-66.5, 13.8) O 190 ASP (111.9, 4.3) O 213 LEU (-52.5, -172.9) O 224 PHE (-42.7, 97.1) O 250 LEU (-37.2, 101.0) O 256 ASN (-42.9, 95.5) O 337 THR (58.6, 91.8) O 362 TYR (172.9, 119.2) O 368 CYS (70.2, 122.1) O 412 GLY (-65.8, -148.9) O 428 SER (-49.6, -75.5) O 478 TRP (-59.7, -165.3) O 479 TYR (82.8, 99.1) O 487 ASN (-178.9, -123.7) O 495 PHE (96.3, 41.7) O 502 VAL (-58.1, -80.8) O 528 GLY (-58.6, -80.8) P 6 PRO (-56.9, -166.7) P 13 LEU (64.8, 122.8) P 48 PRO (-58.1, -74.7) P 49 CYS (-64.8, 78.7) P 122 LEU (-48.4, 167.6) P 146 VAL (-48.3, -76.1) P 147 VAL (63.6, -13.1) P 150 ILE (-61.4, 85.3) P 178 LEU (-46.6, -78.4) P 187 SER (-63.0, 9.3) P 233 GLU (-58.5, -176.5) P 284 ILE (-47.5, -72.2) P 322 LYS (-50.1, 168.7) P 324 PRO (-69.5, -160.2) P 343 ARG (63.9, 140.2) P 356 GLU (-59.2, -78.8) P 373 GLN (62.0, 150.8) P 378 ILE (62.8, 119.0) P 458 VAL (-55.2, -77.3) P 460 PRO (-58.4, -74.2) P 483 ASN (8.2, -40.1) P 484 VAL (60.7, 13.0) P 486 GLU (-144.2, -54.8) P 490 VAL (-162.3, -151.6) http://kinemage.biochem.duke.edu Lovell, Davis, et al. Proteins 50:437 (2003)
Documentos relacionados
Ramachandran Plots
 A 30 PRO (-112.2, 128.0)
A 93 GLN (-46.9, -87.4)
A 188 GLU (29.4, 119.3)
A 191 ARG (-75.5, -132.3)
A 192 VAL (70.9, 123.4)
A 229 GLU (-49.4, 59.3)
A 230 ALA (56.1, -179.6)
A 319 GLU (25.6, 100.7)
A...
A 30 PRO (-112.2, 128.0)
A 93 GLN (-46.9, -87.4)
A 188 GLU (29.4, 119.3)
A 191 ARG (-75.5, -132.3)
A 192 VAL (70.9, 123.4)
A 229 GLU (-49.4, 59.3)
A 230 ALA (56.1, -179.6)
A 319 GLU (25.6, 100.7)
A...
1 pab 1 rdd 1 stj nos 3y par 7y exc 173d 7 oys 81 ll jam 35y abb 3y
 Y26 HDW
C2 HEG
HEJ 595
10 HEN
C110 HEN
PO51 HER
HF 4062
A5 HGF
J1 HGP
HH52 HHH
K44 HHN
190 HHU
HHW 95D
HIL 8992
B5 HJW
777 HJX
1 HKJ
B4 HKO
TT02 HLB
M2 HLL
57 HM
G8 HMF
G1 HMH
1 HMO
HM51 HMS
40 HN
...
Y26 HDW
C2 HEG
HEJ 595
10 HEN
C110 HEN
PO51 HER
HF 4062
A5 HGF
J1 HGP
HH52 HHH
K44 HHN
190 HHU
HHW 95D
HIL 8992
B5 HJW
777 HJX
1 HKJ
B4 HKO
TT02 HLB
M2 HLL
57 HM
G8 HMF
G1 HMH
1 HMO
HM51 HMS
40 HN
...
MolProbity Ramachandran analysis
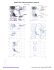 AZ 44 TRP (121.1, 138.3)
BB 24 LYS (-65.6, -165.2)
BB 27 VAL (-154.0, 34.4)
BB 41 LYS (-63.2, -167.9)
BB 52 ILE (-142.9, -107.8)
BB 53 PRO (-92.2, -148.8)
BB 95 TYR (-52.4, 91.0)
BB 113 GLU (131.9,...
AZ 44 TRP (121.1, 138.3)
BB 24 LYS (-65.6, -165.2)
BB 27 VAL (-154.0, 34.4)
BB 41 LYS (-63.2, -167.9)
BB 52 ILE (-142.9, -107.8)
BB 53 PRO (-92.2, -148.8)
BB 95 TYR (-52.4, 91.0)
BB 113 GLU (131.9,...
Polk`s San Francisco (San Francisco County, Calif
 Slmison Walter M (Emily) h2776 Union
Simkins Madyline (wid Clifford E) h349 Pope
" Patti Mrs mgr Trost Apts r3009 Mission
apt 109
" Richd (EHz) mtcemn h286
Cresent av
" Robt (Patti) h3009 Mission
a...
Slmison Walter M (Emily) h2776 Union
Simkins Madyline (wid Clifford E) h349 Pope
" Patti Mrs mgr Trost Apts r3009 Mission
apt 109
" Richd (EHz) mtcemn h286
Cresent av
" Robt (Patti) h3009 Mission
a...
MolProbity Ramachandran analysis
 B 162 SER (-67.9, -82.9)
B 163 SER (42.4, -86.4)
B 179 GLU (0.9, -67.1)
B 213 ARG (-52.8, 97.6)
B 238 LEU (-164.1, -8.0)
B 254 ASP (-23.7, 104.2)
B 255 GLY (-43.7, -120.1)
B 278 ALA (9.5, -102.9)
B...
B 162 SER (-67.9, -82.9)
B 163 SER (42.4, -86.4)
B 179 GLU (0.9, -67.1)
B 213 ARG (-52.8, 97.6)
B 238 LEU (-164.1, -8.0)
B 254 ASP (-23.7, 104.2)
B 255 GLY (-43.7, -120.1)
B 278 ALA (9.5, -102.9)
B...
MolProbity Ramachandran analysis
 O 83 LYS (-54.6, 176.2)
O 116 SER (-55.5, 0.8)
O 143 ALA (48.7, 178.8)
P 6 PRO (-83.8, -43.3)
P 18 PRO (-21.2, -51.1)
P 23 THR (-28.0, 146.8)
O 83 LYS (-54.6, 176.2)
O 116 SER (-55.5, 0.8)
O 143 ALA (48.7, 178.8)
P 6 PRO (-83.8, -43.3)
P 18 PRO (-21.2, -51.1)
P 23 THR (-28.0, 146.8)
MolProbity Ramachandran analysis
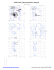 Z 54 PRO (-59.3, -160.9)
Z 55 GLY (-59.4, -111.4)
Z 57 MET (-67.6, -105.2)
Z 98 LYS (-49.4, -5.0)
Z 54 PRO (-59.3, -160.9)
Z 55 GLY (-59.4, -111.4)
Z 57 MET (-67.6, -105.2)
Z 98 LYS (-49.4, -5.0)
MolProbity Ramachandran analysis
 464 SER (-177.1, -0.7)
466 GLY (-10.5, -111.6)
468 PRO (-73.4, -147.7)
469 PHE (-54.0, 79.5)
24 LYS (-45.4, -14.8)
35 GLY (45.2, 7.1)
40 PRO (-84.6, -116.9)
464 SER (-177.1, -0.7)
466 GLY (-10.5, -111.6)
468 PRO (-73.4, -147.7)
469 PHE (-54.0, 79.5)
24 LYS (-45.4, -14.8)
35 GLY (45.2, 7.1)
40 PRO (-84.6, -116.9)
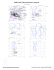 Phi
Phi
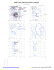 Pre-proline
Pre-proline