Ramachandran Plots
Transcrição
MolProbity Ramachandran analysis 4MEY, model 1 General case 180 Isoleucine and valine 180 K 45 LYS K 15 ASN JASN 718 60D 718 SER SER J 596 1215 GLU JKE LEU ASN C 60 892 GLU CI 941 909 LYS LYS B 13ICLEU G H 188 188 GLU GLU 746 A B 746ALA ALA JD 880 880VAL VAL I D169 148 LYS GLU A 319 319 GLU GLU J 1169 THR B ID 40 595 GLU ALA D 179 LYS C 9 LYS 940 1154 DLYS GLU 1169 ASP THR JCC557 Psi A 229 GLU 0 B Psi 346 TYR ICG346 231TYR PHE E 16 ARG E 80 LEU E 79 GLU D 1208 ASP C 61 SER D 1160 157 SER THR JB 1160 SER G H B A 93 93 93 GLN GLN 9JDLYS 539SER SER 539 B 191 ARG A 191 ARG -180 B 195 ARG I 1155 VAL E 59 ILE 114 VAL K I 59 ILE C 1040 ASP LF 606 606 VAL VAL C 1155 VAL D 1176 VAL J 1176 VAL JD 151 151MET MET LF 488 488 LEU LEU EA 45 LYS 230 ALA Phi C 114 VAL B 192 VAL -180 180 Pre-proline Psi D 825 VAL JD 407 407VAL VAL C 40 GLU 0 180 J 564 VAL J 1209 VAL J 593 ASN D 555 TYR B 193 GLU -180 J 1177 ILE J 582 D 582 ILE ILE 0 C 49 LEU I L 500 ILE ILE F 500 1168GLU GLU JD1168 IC 214 214ASN ASN I 1203 ASP 7 GLU J 560 ASN D ILE AD1155 192 564VAL VAL L 212 ILE D 1177 ILE B A 320 320 ASN ASN C 741 MET MET I 741 JD 462 462ASP ASP J 528 THR L F 307 THR IC 748 748ILE ILE -180 0 180 Glycine 180 Psi 1149 ARG ARG JD 1149 Phi D 906 GLY GLY J 906 D 858 VAL J 522 GLY JD833 833GLU GLU 0 0 D 828 GLY J 1161 GLY G H 29 29 GLU GLU J 587 LEU EK 36 36ASP ASP I 891 GLY JD1190 1190ILE ILE -180 -180 1204LEU LEU IC1204 -180 0 Phi 180 Trans proline 180 834 PRO PRO DJ 834 CI 1181 1181 PRO PRO JDD1185 PRO 1185 PRO 1191 PRO 420 PRO J 1191 PRO JD 420 H PRO G 30 PRO B 30 PRO A 30 PRO D 859 PRO 0 Phi 180 Cis proline 180 JD1150 1150PRO PRO J 859 PRO D 1214 PRO J 1153 D 1153 PRO PRO Psi Psi J 588 PRO D 588 PRO C 110 PRO K 40 PRO 0 -180 FL 490 490 PRO PRO J 1214 PRO 0 E 40 PRO K 37 PRO A 323 PRO E 37 PRO -180 I 110 PRO JD 851 PRO 851 PRO -180 92.5% (6491/7019) of all residues were in favored (98%) regions. 97.6% (6853/7019) of all residues were in allowed (>99.8%) regions. There were 166 outliers (phi, psi): A 30 PRO (-112.2, 128.0) A 93 GLN (-46.9, -87.4) A 188 GLU (29.4, 119.3) A 191 ARG (-75.5, -132.3) A 192 VAL (70.9, 123.4) A 229 GLU (-49.4, 59.3) A 230 ALA (56.1, -179.6) A 319 GLU (25.6, 100.7) A 320 ASN (126.9, 58.9) A 323 PRO (-63.1, -81.9) B 7 GLU (-176.8, -34.0) 0 Phi 180 F 490 PRO (75.9, 7.6) F 500 ILE (-65.3, 86.7) F 606 VAL (87.2, -67.5) G 29 GLU (-77.1, -81.6) H H H I 110 PRO (77.9, -175.9) I 114 VAL (-69.3, -87.2) I 169 LYS (59.3, 103.1) I 214 ASN (110.6, 24.7) 29 GLU (-75.5, -81.6) 30 PRO (-108.8, 143.1) 93 GLN (-48.7, -88.0) B 320 ASN (125.1, 58.3) C 9 LYS (49.5, 82.4) C 40 GLU (46.8, -159.8) C 49 LEU (58.2, -31.9) I 346 TYR (39.6, -59.9) I 741 MET (29.0, 50.0) I 746 ALA (91.9, 120.3) I 748 ILE (144.0, 161.4) C 61 SER (34.2, -107.7) C 110 PRO (-154.9, 17.1) C 114 VAL (-67.7, -164.6) I 891 GLY (-67.7, -129.0) I 941 LYS (64.8, 126.2) I 1155 VAL (-40.2, -66.1) I 1181 PRO (-109.0, 163.7) I 1203 ASP (90.3, 18.5) I 1204 LEU (73.3, -178.6) J 151 MET (110.7, -161.5) J 407 VAL (81.8, -11.5) J 420 PRO (-123.0, 143.2) J 462 ASP (-68.7, 18.1) C 940 GLU (-60.2, 78.0) C 1040 ASP (174.3, -74.2) C 1154 ASP (-56.8, 78.1) C 1155 VAL (44.6, -69.1) J 522 GLY (166.5, 28.5) J 528 THR (-48.4, 11.2) J 539 SER (-46.9, -103.0) J 557 LYS (-61.3, 77.2) C 1181 PRO (-111.2, 160.8) C 1204 LEU (73.2, -177.5) D 148 GLU (65.2, 103.9) J 560 ASN (-171.0, -69.3) J 564 VAL (72.6, 43.5) J 582 ILE (-70.8, 61.4) D 151 MET (112.4, -162.9) D 179 LYS (-62.3, 90.5) D 407 VAL (81.1, -13.6) D 420 PRO (-121.4, 145.1) J 587 LEU (55.1, -111.7) J 588 PRO (47.8, 54.7) J 593 ASN (71.0, -135.1) J 596 LEU (61.0, 142.1) D 462 ASP (-66.3, 17.9) D 539 SER (-45.7, -100.0) D 555 TYR (84.1, -142.4) D 564 VAL (76.5, 119.7) J 718 SER (88.9, 149.1) J 833 GLU (72.9, 19.4) J 834 PRO (-106.9, 175.8) J 851 PRO (62.8, -176.3) D 582 ILE (-72.6, 57.3) D 588 PRO (-151.3, 50.1) D 595 ALA (55.8, 91.1) -180 G 30 PRO (-108.5, 142.8) G 93 GLN (-48.8, -87.6) G 188 GLU (30.4, 121.2) G 231 PHE (42.5, -60.1) H 188 GLU (30.3, 121.3) I 9 LYS (-61.1, -97.5) I 40 GLU (53.8, 91.1) B 192 VAL (75.3, -175.8) B 193 GLU (-65.7, -168.2) B 195 ARG (172.4, -39.4) B 319 GLU (24.4, 96.9) D 718 SER (87.7, 146.8) D 825 VAL (62.7, -0.9) D 828 GLY (-178.5, -42.7) D 833 GLU (76.6, 17.8) -180 180 E 80 LEU (46.0, -81.0) F 307 THR (-51.5, -2.0) F 488 LEU (153.5, -165.7) B 157 THR (-38.6, -77.1) B 188 GLU (30.6, 119.0) B 191 ARG (-74.6, -124.0) C 214 ASN (110.6, 24.0) C 346 TYR (39.6, -56.5) C 741 MET (28.4, 50.6) C 746 ALA (92.9, 119.0) Phi E 45 LYS (52.3, -176.1) E 59 ILE (-78.8, -80.2) E 60 ASN (64.5, 141.4) E 79 GLU (53.7, -91.1) B 13 LEU (66.4, 122.8) B 30 PRO (-113.6, 133.2) B 93 GLN (-46.8, -87.6) C 748 ILE (144.7, 160.0) C 892 GLU (67.4, 137.2) C 909 LYS (62.3, 129.5) 0 J 859 PRO (52.1, 127.2) J 880 VAL (73.2, 154.9) J 906 GLY (-33.6, 92.4) J 1149 ARG (-80.2, 83.5) J 1150 PRO (-27.3, 133.4) J 1153 PRO (52.5, 103.3) J 1160 SER (-44.6, -80.1) D 834 PRO (-112.5, 173.3) D 851 PRO (62.6, -177.7) D 858 VAL (-97.9, 32.4) J 1161 GLY (58.2, -75.4) J 1168 GLU (164.8, 32.5) J 1169 THR (-36.9, 98.2) D 859 PRO (-129.5, 121.8) D 880 VAL (74.1, 153.4) D 906 GLY (-34.3, 93.8) D 1149 ARG (-80.7, 86.6) J 1176 VAL (60.4, -146.5) J 1177 ILE (58.1, 84.6) J 1185 PRO (-135.4, 151.3) J 1190 ILE (-89.6, -154.8) D 1150 PRO (-25.9, 133.0) D 1153 PRO (51.3, 102.4) D 1155 ILE (68.2, 127.9) J 1191 PRO (-132.2, 144.9) J 1209 VAL (-162.0, 10.1) J 1214 PRO (49.3, -1.3) D 1160 SER (-57.9, -77.0) D 1168 GLU (167.4, 34.3) D 1169 THR (-37.9, 78.9) D 1176 VAL (62.2, -129.6) J 1215 GLU (64.1, 144.5) K 15 ASN (92.1, 158.8) K 36 ASP (35.1, -131.0) K 37 PRO (55.0, -71.2) D 1177 ILE (83.0, 102.8) D 1185 PRO (-135.4, 150.2) D 1190 ILE (-88.8, -155.2) K 40 PRO (-148.9, 6.6) K 45 LYS (52.1, 179.5) K 59 ILE (-78.4, -88.5) D 1191 PRO (-132.5, 147.4) D 1208 ASP (71.3, -98.1) D 1214 PRO (-0.2, 112.8) E 16 ARG (46.7, -73.2) K 60 ASN (61.8, 147.3) L 212 ILE (63.3, 112.3) L 307 THR (-51.5, -1.8) L 488 LEU (151.8, -164.2) E 36 ASP (32.7, -129.9) E 37 PRO (61.7, -143.6) E 40 PRO (142.9, 5.4) L 490 PRO (76.6, 7.4) L 500 ILE (-65.3, 89.3) L 606 VAL (84.9, -67.2) http://kinemage.biochem.duke.edu Lovell, Davis, et al. Proteins 50:437 (2003)
Documentos relacionados
MolProbity Ramachandran analysis
 O 83 LYS (-54.6, 176.2)
O 116 SER (-55.5, 0.8)
O 143 ALA (48.7, 178.8)
P 6 PRO (-83.8, -43.3)
P 18 PRO (-21.2, -51.1)
P 23 THR (-28.0, 146.8)
O 83 LYS (-54.6, 176.2)
O 116 SER (-55.5, 0.8)
O 143 ALA (48.7, 178.8)
P 6 PRO (-83.8, -43.3)
P 18 PRO (-21.2, -51.1)
P 23 THR (-28.0, 146.8)
MolProbity Ramachandran analysis
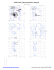 Z 54 PRO (-59.3, -160.9)
Z 55 GLY (-59.4, -111.4)
Z 57 MET (-67.6, -105.2)
Z 98 LYS (-49.4, -5.0)
Z 54 PRO (-59.3, -160.9)
Z 55 GLY (-59.4, -111.4)
Z 57 MET (-67.6, -105.2)
Z 98 LYS (-49.4, -5.0)
MolProbity Ramachandran analysis
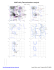 H 120 GLY (57.4, 86.8)
I 23 PRO (-61.5, 75.4)
I 26 GLY (-54.3, 68.9)
I 39 PHE (-167.6, 23.9)
I 41 ARG (167.2, -39.7)
I 53 GLU (-85.8, -112.7)
I 55 VAL (67.4, 116.3)
I 58 VAL (57.1, -67.3)
I 103 PHE...
H 120 GLY (57.4, 86.8)
I 23 PRO (-61.5, 75.4)
I 26 GLY (-54.3, 68.9)
I 39 PHE (-167.6, 23.9)
I 41 ARG (167.2, -39.7)
I 53 GLU (-85.8, -112.7)
I 55 VAL (67.4, 116.3)
I 58 VAL (57.1, -67.3)
I 103 PHE...
MolProbity Ramachandran analysis
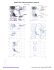 AZ 44 TRP (121.1, 138.3)
BB 24 LYS (-65.6, -165.2)
BB 27 VAL (-154.0, 34.4)
BB 41 LYS (-63.2, -167.9)
BB 52 ILE (-142.9, -107.8)
BB 53 PRO (-92.2, -148.8)
BB 95 TYR (-52.4, 91.0)
BB 113 GLU (131.9,...
AZ 44 TRP (121.1, 138.3)
BB 24 LYS (-65.6, -165.2)
BB 27 VAL (-154.0, 34.4)
BB 41 LYS (-63.2, -167.9)
BB 52 ILE (-142.9, -107.8)
BB 53 PRO (-92.2, -148.8)
BB 95 TYR (-52.4, 91.0)
BB 113 GLU (131.9,...
MolProbity Ramachandran analysis
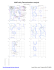 205 THR (154.1, 65.9)
210 VAL (160.1, -177.3)
211 ALA (-137.1, -164.6)
212 PRO (-112.7, -107.1)
213 THR (118.1, -175.2)
215 CYS (88.8, 139.1)
2 SER (80.8, -78.4)
3 ALA (161.4, -103.5)
4 LEU (161.6,...
205 THR (154.1, 65.9)
210 VAL (160.1, -177.3)
211 ALA (-137.1, -164.6)
212 PRO (-112.7, -107.1)
213 THR (118.1, -175.2)
215 CYS (88.8, 139.1)
2 SER (80.8, -78.4)
3 ALA (161.4, -103.5)
4 LEU (161.6,...
MolProbity Ramachandran analysis
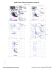 A 94 ILE (-148.3, -91.7)
A 121 HIS (79.1, 126.5)
A 199 PRO (-38.6, 103.6)
A 265 ASP (160.6, 146.6)
A 267 PRO (-40.3, 160.8)
A 292 GLY (176.9, -81.6)
A 335 ALA (-60.7, -171.9)
A 337 LEU (-54.9, 96.6...
A 94 ILE (-148.3, -91.7)
A 121 HIS (79.1, 126.5)
A 199 PRO (-38.6, 103.6)
A 265 ASP (160.6, 146.6)
A 267 PRO (-40.3, 160.8)
A 292 GLY (176.9, -81.6)
A 335 ALA (-60.7, -171.9)
A 337 LEU (-54.9, 96.6...
MolProbity Ramachandran analysis
 464 SER (-177.1, -0.7)
466 GLY (-10.5, -111.6)
468 PRO (-73.4, -147.7)
469 PHE (-54.0, 79.5)
24 LYS (-45.4, -14.8)
35 GLY (45.2, 7.1)
40 PRO (-84.6, -116.9)
464 SER (-177.1, -0.7)
466 GLY (-10.5, -111.6)
468 PRO (-73.4, -147.7)
469 PHE (-54.0, 79.5)
24 LYS (-45.4, -14.8)
35 GLY (45.2, 7.1)
40 PRO (-84.6, -116.9)
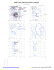 Pre-proline
Pre-proline
 BA1491
SER
AA1596
2112ASP
ALA
B 1302 ALA
BA1491
SER
AA1596
2112ASP
ALA
B 1302 ALA
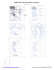 Pre-proline
Pre-proline