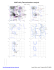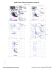MolProbity Ramachandran analysis
Transcrição
MolProbity Ramachandran analysis 1IYJ, model 1 General case 180 Psi A ASP 99 PHE ASP BB 2406 DC 3040 3040 ASP ASP B 2954 2954 GLN GLN D D 2944 2944 SER B SER B D 2738 2738 LEU LEU BBD LYS D 2721 3008 3008 SER SER D B 2501 GLU D 2910 2910 GLU GLU B B 2686 2686 ALA ALA D C 55 GLN B 2942 SER 2942SER SER A 55 GLN DD2912 2912 B SER D B 2694 2694 ALA B D 2512 2512 ALA ALA Isoleucine and valine 180 B DD2764 2764 THR THR D2464 2460ALA SER B 2460 SER DB 2930 2930 GLU GLU A Psi 2752 VAL VAL DB 2752 8 VAL D3114 3099ASP PHE 3099 PHE DB A C 44 ASP B 2998 2998 SER BD 3038 3038 ASN ASN D D ASN B 3095 3095 ASN D B D 2429 2611 2611 GLU THR THR B 2429 GLU A 51 ASP C 51 ASP DD2988 2988 LEU B LEU 2710 ALA CB BB 10 2992 LEU ALA 2710 2992 ALA ALA DD 2407 ASN 2407 ASN DBD SER B2672 3092 2415 ALA GLN 3092 ALA BD D 3088 3101ASN B 3088 ASN LYS D 2415 GLN B 2564 MET D 2564 D 2985 MET PHE 0 B 2945 LYS A C 39 TRP 2610 2610ASP ASP B 3114BDASP 48 VAL VAL CA 48 0 D 2480 2480 ILE ILE B D 2945 LYS BD 2941 2941VAL VAL AC 57 57ARG ARG B D2970 2507 2507SER LYS LYS B D 2970 SER B 2779 2779 LYS LYS D B DB 2573 ALA D2573 2909 2909ALA ARG ARGD B 2490 2490 PHE D 2893 ARG BA 2893 C 53 53 ARG SER SER D 2465 B CYS D B 2447 THR D 3039 GLU 2907LYS LYS B 2907 B DD2647 2647 ASP ASP 41 ASP ASP CA 41 D B 2592 2592 VAL D B 2605 2605 ILE ILE BD 2737 2737 SER SER C 47 47 ASN ASN A B D 2946 2946 ASN ASN -180 -180 0 180 Phi Pre-proline 180 B D 3035 3035 ILE ILE D 2953 2953 ILE ILE B -180 -180 0 Phi 180 Glycine 180 BD 2448 2448LEU LEU Psi B 2474 GLY Psi D B 3016 ALA D 2919 ARG D B 2466 SER 0 0 B D 2950 2950 TRP TRP D B 2685 GLY -180 -180 -180 0 Phi 180 Trans proline 180 A C 7 PRO 0 0 Phi 180 Phi 180 Cis proline 180 D B 2979 2979 PRO B D 2519 2519 PRO PRO B 3063 PRO D B 2462 PRO Psi -180 Psi 0 B 2569 2569 PRO D BD2697 2697PRO PRO D B 2991 PRO B D 2467 2467 PRO PRO B D 2692 2692 PRO PRO -180 -180 -180 65.0% (814/1252) of all residues were in favored (98%) regions. 86.7% (1086/1252) of all residues were in allowed (>99.8%) regions. There were 166 outliers (phi, psi): A A A 7 PRO (-109.8, 170.4) 8 VAL (-59.1, 99.6) 9 ASP (-53.4, 89.4) A 39 TRP (-175.3, -50.9) A 41 ASP (-20.2, -112.0) A 44 ASP (-68.4, 38.2) A 47 ASN (-59.5, -156.2) -180 0 C 39 TRP (-174.3, -50.2) C 41 ASP (-22.6, -113.8) C 44 ASP (-67.6, 38.6) D 2429 GLU (-52.5, 15.6) D 2447 THR (-81.1, -93.9) D 2448 LEU (46.8, 111.8) B 2415 GLN (-34.9, -24.0) B 2429 GLU (-55.1, 10.3) B 2447 THR (-80.6, -93.2) D 2460 SER (-45.1, 167.4) D 2462 PRO (-55.3, 100.2) D 2464 ALA (-49.8, 169.2) B 2448 LEU (44.7, 113.8) B 2460 SER (-45.1, 166.7) B 2462 PRO (-54.1, 100.1) B 2465 CYS (-54.9, -91.4) D 2465 CYS (-55.9, -91.0) D 2466 SER (151.7, 31.7) D 2467 PRO (55.6, -80.9) D 2480 ILE (-51.9, -6.8) B 2466 SER (152.1, 31.7) B 2467 PRO (55.4, -80.5) B 2474 GLY (73.4, 97.0) B 2480 ILE (-52.2, -7.6) D 2490 PHE (64.3, -79.1) D 2501 GLU (138.8, 119.9) D 2507 LYS (-15.3, -62.2) D 2512 ALA (-14.5, 133.3) D 2519 PRO (-35.6, 159.0) D 2564 MET (-29.4, -39.2) D 2569 PRO (-42.8, -10.7) D 2573 ALA (2.6, -77.8) D 2592 VAL (-47.1, -69.2) D 2605 ILE (-50.9, -75.4) D 2610 ASP (179.3, 39.6) B 2573 ALA (1.8, -77.7) B 2592 VAL (-45.1, -70.7) B 2605 ILE (-48.4, -79.0) D 2611 THR (-51.9, 15.2) D 2647 ASP (-58.5, -100.3) D 2672 SER (-43.4, -21.4) B 2610 ASP (174.9, 39.1) B 2611 THR (-51.7, 15.8) B 2647 ASP (-59.1, -100.2) B 2685 GLY (-45.2, -80.0) D 2685 GLY (-46.7, -78.4) D 2686 ALA (45.7, 110.8) D 2692 PRO (-90.4, -110.6) D 2694 ALA (98.9, 94.9) B 2686 ALA (47.2, 112.1) B 2692 PRO (-90.8, -109.6) B 2694 ALA (100.0, 95.4) D 2697 PRO (-19.4, -48.9) D 2710 ALA (-49.1, -7.0) D 2721 LYS (166.5, 134.1) B 2697 PRO (-22.8, -43.0) B 2710 ALA (-46.3, -13.0) B 2721 LYS (167.1, 135.5) B 2737 SER (-72.5, -129.8) D 2737 SER (-70.3, -130.1) D 2738 LEU (65.8, 140.6) D 2752 VAL (-18.7, 104.9) D 2764 THR (-52.8, 174.9) B 2738 LEU (64.8, 140.0) B 2752 VAL (-15.2, 105.3) B 2764 THR (-52.5, 174.0) B 2779 LYS (-45.7, -72.8) D 2779 LYS (-47.1, -74.1) D 2893 ARG (-53.6, -80.5) D 2907 LYS (-55.8, -96.8) D 2909 ARG (6.1, -78.9) B 2893 ARG (-52.5, -82.2) B 2907 LYS (-58.2, -98.8) B 2909 ARG (6.6, -78.3) D 2910 GLU (92.6, 114.9) D 2912 SER (32.3, 97.1) D 2919 ARG (37.7, 35.0) B 2910 GLU (91.5, 113.3) B 2912 SER (32.1, 96.3) B 2930 GLU (-30.1, 155.9) B 2941 VAL (146.5, -32.1) D 2930 GLU (-34.2, 155.4) D 2941 VAL (148.0, -32.0) D 2942 SER (37.3, 102.4) D 2944 SER (50.6, 143.2) B 2942 SER (36.0, 104.6) B 2944 SER (49.9, 142.6) B 2945 LYS (-179.9, -30.0) D 2945 LYS (178.9, -29.5) D 2946 ASN (-55.6, -173.2) D 2950 TRP (-165.6, -23.7) B 2946 ASN (-55.5, -172.5) B 2950 TRP (-166.0, -23.4) B 2953 ILE (-78.5, -169.6) B 2954 GLN (-176.3, 84.4) 180 C 47 ASN (-59.0, -153.9) C 48 VAL (-155.1, 34.0) C 51 ASP (-38.7, -0.4) C 53 SER (-44.2, -85.7) C 55 GLN (103.9, 105.1) C 57 ARG (105.4, -55.1) D 2407 ASN (-46.4, -14.0) D 2415 GLN (-33.6, -31.4) A 57 ARG (101.9, -53.6) B 2406 PHE (-63.1, 84.7) B 2407 ASN (-38.9, -15.3) B 2512 ALA (-14.8, 134.1) B 2519 PRO (-35.7, 160.1) B 2564 MET (-30.2, -38.7) B 2569 PRO (-40.9, -9.4) Phi B 3114 ASP (147.7, 35.5) C 7 PRO (-110.0, 169.9) C 9 ASP (-54.9, 86.4) C 10 LEU (-48.4, -11.3) A 48 VAL (-153.4, 34.7) A 51 ASP (-41.1, 1.2) A 53 SER (-46.1, -85.1) A 55 GLN (103.6, 102.6) B 2490 PHE (66.5, -79.0) B 2501 GLU (139.8, 118.4) B 2507 LYS (-16.2, -62.4) 0 D 2953 ILE (-77.4, -168.8) D 2954 GLN (-178.5, 81.9) D 2970 SER (-18.4, -64.1) D 2979 PRO (-40.7, 168.3) B 2970 SER (-19.1, -64.2) B 2979 PRO (-38.1, 166.1) B 2988 LEU (-52.1, -3.6) D 2985 PHE (-29.0, -45.0) D 2988 LEU (-52.8, -2.2) D 2991 PRO (-48.1, -71.1) B 2991 PRO (-47.4, -72.5) B 2992 ALA (-42.9, -11.5) B 2998 SER (33.3, 32.3) B 3008 SER (173.4, 133.6) D 2992 ALA (-43.3, -13.4) D 2998 SER (31.2, 31.4) D 3008 SER (175.5, 133.8) D 3016 ALA (102.7, 53.2) B 3016 ALA (103.2, 53.6) B 3035 ILE (-87.0, -159.5) B 3038 ASN (-44.4, 31.9) D 3035 ILE (-86.6, -159.3) D 3038 ASN (-42.2, 29.8) D 3039 GLU (63.7, -95.1) B 3040 ASP (-60.2, 82.7) B 3063 PRO (-39.3, 149.8) B 3088 ASN (-39.2, -27.7) B 3092 ALA (-38.1, -24.6) D 3040 ASP (-60.8, 82.6) D 3088 ASN (-36.8, -28.7) D 3092 ALA (-35.0, -25.2) D 3095 ASN (-65.6, 18.4) B 3095 ASN (-64.8, 16.8) B 3099 PHE (176.7, 59.6) B 3101 LYS (-30.8, -28.2) D 3099 PHE (176.5, 61.3) D 3101 LYS (-31.6, -27.4) D 3114 ASP (171.1, 56.6) http://kinemage.biochem.duke.edu Lovell, Davis, et al. Proteins 50:437 (2003)
Documentos relacionados
MolProbity Ramachandran analysis
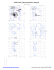 Q 274 ILE (-22.8, -54.8)
R 2 GLU (-25.5, -50.1)
R 233 SER (47.8, 90.0)
R 242 LEU (-65.2, -171.2)
R 246 GLN (-18.7, 73.5)
R 253 GLU (-40.7, -26.2)
R 274 ILE (-23.0, -54.6)
S 2 GLU (-25.5, -50.3)
S 2...
Q 274 ILE (-22.8, -54.8)
R 2 GLU (-25.5, -50.1)
R 233 SER (47.8, 90.0)
R 242 LEU (-65.2, -171.2)
R 246 GLN (-18.7, 73.5)
R 253 GLU (-40.7, -26.2)
R 274 ILE (-23.0, -54.6)
S 2 GLU (-25.5, -50.3)
S 2...
Ramachandran Plots
 A 30 PRO (-112.2, 128.0)
A 93 GLN (-46.9, -87.4)
A 188 GLU (29.4, 119.3)
A 191 ARG (-75.5, -132.3)
A 192 VAL (70.9, 123.4)
A 229 GLU (-49.4, 59.3)
A 230 ALA (56.1, -179.6)
A 319 GLU (25.6, 100.7)
A...
A 30 PRO (-112.2, 128.0)
A 93 GLN (-46.9, -87.4)
A 188 GLU (29.4, 119.3)
A 191 ARG (-75.5, -132.3)
A 192 VAL (70.9, 123.4)
A 229 GLU (-49.4, 59.3)
A 230 ALA (56.1, -179.6)
A 319 GLU (25.6, 100.7)
A...
MolProbity Ramachandran analysis
 3 ALA (87.5, -98.2)
4 ALA (171.2, 16.2)
6 GLY (-174.8, -58.2)
7 ASP (9.6, -102.1)
8 HIS (49.8, -18.1)
39 SER (-61.4, -124.3)
40 GLY (65.7, 129.7)
49 PRO (-59.4, 53.6)
8 ILE (78.9, 10.3)
10 ASN (88....
3 ALA (87.5, -98.2)
4 ALA (171.2, 16.2)
6 GLY (-174.8, -58.2)
7 ASP (9.6, -102.1)
8 HIS (49.8, -18.1)
39 SER (-61.4, -124.3)
40 GLY (65.7, 129.7)
49 PRO (-59.4, 53.6)
8 ILE (78.9, 10.3)
10 ASN (88....
Ramachandran Plots
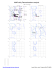 B 6 ALA (53.1, -88.5)
B 12 GLU (-66.3, -157.7)
B 13 ARG (38.0, -152.0)
B 19 ILE (-53.9, -79.5)
B 20 LYS (-28.0, -42.9)
B 37 LYS (-47.4, 90.3)
B 61 LEU (-59.1, -78.1)
B 71 GLN (-58.2, -86.0)
B 72 AS...
B 6 ALA (53.1, -88.5)
B 12 GLU (-66.3, -157.7)
B 13 ARG (38.0, -152.0)
B 19 ILE (-53.9, -79.5)
B 20 LYS (-28.0, -42.9)
B 37 LYS (-47.4, 90.3)
B 61 LEU (-59.1, -78.1)
B 71 GLN (-58.2, -86.0)
B 72 AS...
MolProbity Ramachandran analysis
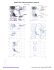 AZ 44 TRP (121.1, 138.3)
BB 24 LYS (-65.6, -165.2)
BB 27 VAL (-154.0, 34.4)
BB 41 LYS (-63.2, -167.9)
BB 52 ILE (-142.9, -107.8)
BB 53 PRO (-92.2, -148.8)
BB 95 TYR (-52.4, 91.0)
BB 113 GLU (131.9,...
AZ 44 TRP (121.1, 138.3)
BB 24 LYS (-65.6, -165.2)
BB 27 VAL (-154.0, 34.4)
BB 41 LYS (-63.2, -167.9)
BB 52 ILE (-142.9, -107.8)
BB 53 PRO (-92.2, -148.8)
BB 95 TYR (-52.4, 91.0)
BB 113 GLU (131.9,...
MolProbity Ramachandran analysis
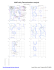 205 THR (154.1, 65.9)
210 VAL (160.1, -177.3)
211 ALA (-137.1, -164.6)
212 PRO (-112.7, -107.1)
213 THR (118.1, -175.2)
215 CYS (88.8, 139.1)
2 SER (80.8, -78.4)
3 ALA (161.4, -103.5)
4 LEU (161.6,...
205 THR (154.1, 65.9)
210 VAL (160.1, -177.3)
211 ALA (-137.1, -164.6)
212 PRO (-112.7, -107.1)
213 THR (118.1, -175.2)
215 CYS (88.8, 139.1)
2 SER (80.8, -78.4)
3 ALA (161.4, -103.5)
4 LEU (161.6,...
MolProbity Ramachandran analysis
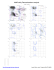 B 426 LYS (-61.4, -84.8)
B 427 ASP (-7.7, -71.3)
B 429 PHE (-54.8, 3.1)
B 509 ALA (82.5, 77.3)
B 510 LYS (-98.6, 59.8)
B 574 SER (-179.4, 52.3)
B 644 GLU (-173.6, -63.8)
B 645 SER (102.2, -104.5)
B...
B 426 LYS (-61.4, -84.8)
B 427 ASP (-7.7, -71.3)
B 429 PHE (-54.8, 3.1)
B 509 ALA (82.5, 77.3)
B 510 LYS (-98.6, 59.8)
B 574 SER (-179.4, 52.3)
B 644 GLU (-173.6, -63.8)
B 645 SER (102.2, -104.5)
B...
MolProbity Ramachandran analysis
 464 SER (-177.1, -0.7)
466 GLY (-10.5, -111.6)
468 PRO (-73.4, -147.7)
469 PHE (-54.0, 79.5)
24 LYS (-45.4, -14.8)
35 GLY (45.2, 7.1)
40 PRO (-84.6, -116.9)
464 SER (-177.1, -0.7)
466 GLY (-10.5, -111.6)
468 PRO (-73.4, -147.7)
469 PHE (-54.0, 79.5)
24 LYS (-45.4, -14.8)
35 GLY (45.2, 7.1)
40 PRO (-84.6, -116.9)
MolProbity Ramachandran analysis
 O 83 LYS (-54.6, 176.2)
O 116 SER (-55.5, 0.8)
O 143 ALA (48.7, 178.8)
P 6 PRO (-83.8, -43.3)
P 18 PRO (-21.2, -51.1)
P 23 THR (-28.0, 146.8)
O 83 LYS (-54.6, 176.2)
O 116 SER (-55.5, 0.8)
O 143 ALA (48.7, 178.8)
P 6 PRO (-83.8, -43.3)
P 18 PRO (-21.2, -51.1)
P 23 THR (-28.0, 146.8)
Ramachandran Plots
 75 THR (57.9, 107.4)
76 GLU (-176.0, -40.0)
78 ILE (-159.0, -42.6)
79 SER (60.6, 111.1)
84 GLN (59.3, 99.0)
85 VAL (59.3, 88.4)
89 LYS (-175.7, -40.1)
95 PRO (-53.5, -172.8)
96 LYS (-177.3, -39.6)
...
75 THR (57.9, 107.4)
76 GLU (-176.0, -40.0)
78 ILE (-159.0, -42.6)
79 SER (60.6, 111.1)
84 GLN (59.3, 99.0)
85 VAL (59.3, 88.4)
89 LYS (-175.7, -40.1)
95 PRO (-53.5, -172.8)
96 LYS (-177.3, -39.6)
...
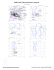 Phi
Phi
 BA1491
SER
AA1596
2112ASP
ALA
B 1302 ALA
BA1491
SER
AA1596
2112ASP
ALA
B 1302 ALA
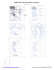 Pre-proline
Pre-proline